Complex II ambiguities: Difference between revisions
No edit summary |
No edit summary |
||
| Line 5: | Line 5: | ||
}} | }} | ||
== FADH<sub>2</sub> and S-pathway == | == FADH<sub>2</sub> and S-pathway == | ||
:::: [[File:Arnold, Finley 2022 CORRECTION.png| | :::: [[File:Arnold, Finley 2022 CORRECTION.png|600px|link=Arnold 2022 J Biol Chem]] | ||
<br> | <br> | ||
:::: [[File:Chen 2022 Am J Physiol Cell Physiol CORRECTION.png| | :::: [[File:Chen 2022 Am J Physiol Cell Physiol CORRECTION.png|500px|link=Chen 2022 Am J Physiol Cell Physiol]] | ||
<br> | <br> | ||
:::: [[File:Turton 2022 Int J Mol Sci CORRECTION.png| | :::: [[File:Turton 2022 Int J Mol Sci CORRECTION.png|500px|link=Turton 2022 Int J Mol Sci]] | ||
<br> | <br> | ||
:::: [[File:Ahmad 2022 StatPearls CORRECTION.png| | :::: [[File:Ahmad 2022 StatPearls CORRECTION.png|400px|link=Ahmad 2022 StatPearls Publishing]] | ||
<br> | <br> | ||
:::: [[File:Martinez-Reyes, Chandel 2020 CORRECTION.png| | :::: [[File:Martinez-Reyes, Chandel 2020 CORRECTION.png|600px|link=Martinez-Reyes 2020 Nat Commun]] | ||
<br> | <br> | ||
:::: [[File:Morelli 2019 Open Biol CORRECTION.png| | :::: [[File:Morelli 2019 Open Biol CORRECTION.png|500px|left|link=Morelli 2019 Open Biol]] | ||
<br> | <br> | ||
:::: [[File:Yepez 2018 PLOS One Fig1B.jpg| | :::: [[File:Yepez 2018 PLOS One Fig1B.jpg|300px|left|link=Yepez 2018 PLOS One]] | ||
<br> | <br> | ||
:::: [[File:Chowdhury 2018 Oxid Med Cell Longev CORRECTION.png| | :::: [[File:Chowdhury 2018 Oxid Med Cell Longev CORRECTION.png|400px|left|link=Chowdhury 2018 Oxid Med Cell Longev]] | ||
<br> | <br> | ||
:::: [[File:Zhang 2018 Mil Med Res CORRECTION.png| | :::: [[File:Zhang 2018 Mil Med Res CORRECTION.png|400px|left|link=Zhang 2018 Mil Med Res]] | ||
<br> | <br> | ||
:::: [[File:DeBerardinis, Chandel 2016 CORRECTION.png| | :::: [[File:DeBerardinis, Chandel 2016 CORRECTION.png|600px|link=DeBerardinis 2016 Sci Adv]] | ||
<br> | <br> | ||
| Line 38: | Line 38: | ||
<br> | <br> | ||
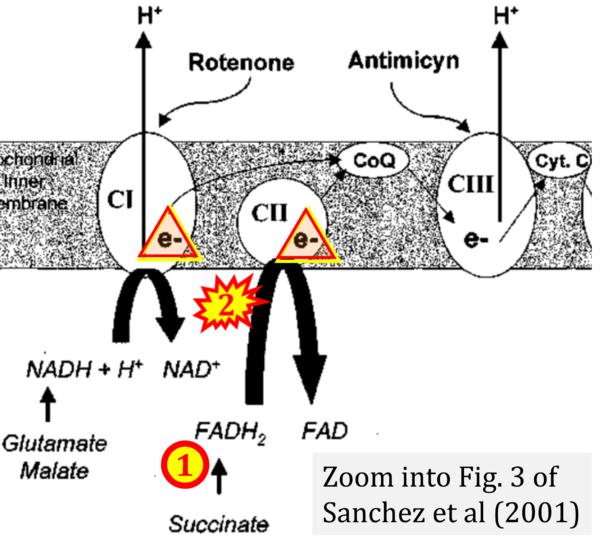
:::: [[File:Sanchez et al 2001 CORRECTION.png| | :::: [[File:Sanchez et al 2001 CORRECTION.png|600px|link=Sanchez 2001 Br J Pharmacol]] | ||
<br> | <br> | ||
Revision as of 08:53, 27 February 2023
Description
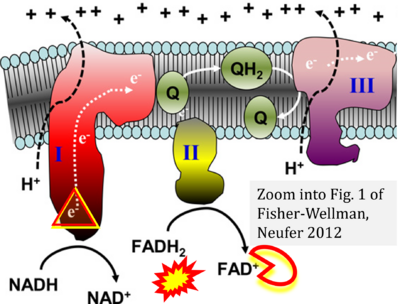
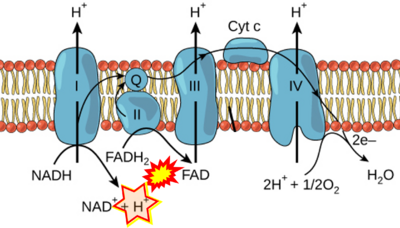
The Succinate pathway is represented in the literature in some cases with a surprising confusion which warrants an analysis of Complex II ambiguities. The CII ambiguity has its roots in the narrative that reduced coenzymes (NADH and FADH2) feed electrons from the Krebs cycle into the membrane-bound electron transfer system. In corresponding ambiguous graphical representations, CII in the canonical ('forward') Krebs cycle is shown to reduce FAD to FADH2 (correct), yet CII in the membrane-bound electron transfer system is paradoxically represented as the site of oxidation of FADH2 to FAD. For clarification: "The substrate of CII is succinate, which is oxidized forming fumarate while reducing flavin adenine dinucleotide FAD to FADH2, with further electron transfer to the quinone pool. Whereas reduced NADH is a substrate of Complex I linked to dehydrogenases of the TCA cycle and mt-matrix upstream of CI, reduced FADH2 is a product of Complex II with downstream electron flow from CII to Q" (quote from Gnaiger 2020).
Abbreviation: CII ambiguities
Reference: Gnaiger E (2020) Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis. 5th ed. Bioenerg Commun 2020.2. https://doi.org/10.26124/bec:2020-0002
FADH2 and S-pathway
- FADH2 appears in several publications as the substrate of CII in the electron transfer system. This commonly found error requires correction. For clarification, see Gnaiger (2020) page 48.
- FADH2→CII misconceptions: references and links
- Arnold PK, Finley LWS (2022) Regulation and function of the mammalian tricarboxylic acid cycle. J Biol Chem 299:102838. - »Bioblast link«
- Chen CL, Zhang L, Jin Z, Kasumov T, Chen YR (2022) Mitochondrial redox regulation and myocardial ischemia-reperfusion injury. Am J Physiol Cell Physiol 322:C12-23. - »Bioblast link«
- Turton N, Cufflin N, Dewsbury M, Fitzpatrick O, Islam R, Watler LL, McPartland C, Whitelaw S, Connor C, Morris C, Fang J, Gartland O, Holt L, Hargreaves IP (2022) The biochemical assessment of mitochondrial respiratory chain disorders. Int J Mol Sci 23:7487. - »Bioblast link«
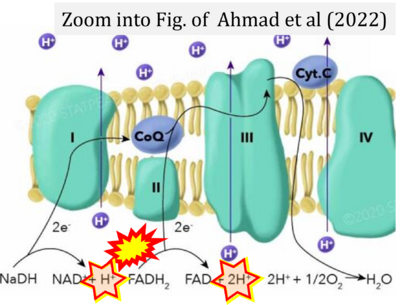
- Ahmad M, Wolberg A, Kahwaji CI (2022) Biochemistry, electron transport chain. StatPearls Publishing StatPearls [Internet]. Treasure Island (FL) - »Bioblast link«
- Martínez-Reyes I, Chandel NS (2020) Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 11:102. - »Bioblast link«
- Morelli AM, Ravera S, Calzia D, Panfoli I (2019) An update of the chemiosmotic theory as suggested by possible proton currents inside the coupling membrane. Open Biol 9:180221. - »Bioblast link«
- Yépez VA, Kremer LS, Iuso A, Gusic M, Kopajtich R, Koňaříková E, Nadel A, Wachutka L, Prokisch H, Gagneur J (2018) OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. PLOS ONE 13:e0199938. - »Bioblast link«
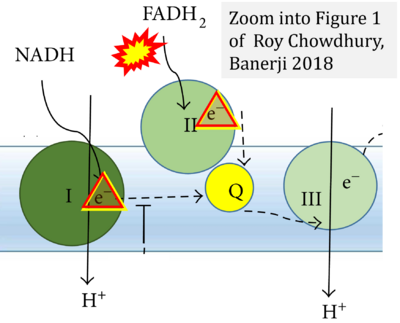
- Roy Chowdhury S, Banerji V (2018) Targeting mitochondrial bioenergetics as a therapeutic strategy for chronic lymphocytic leukemia. Oxid Med Cell Longev 2018:2426712. - »Bioblast link«
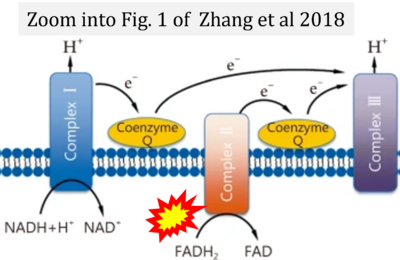
- Zhang H, Feng YW, Yao YM (2018) Potential therapy strategy: targeting mitochondrial dysfunction in sepsis. Mil Med Res 5:41. - »Bioblast link«
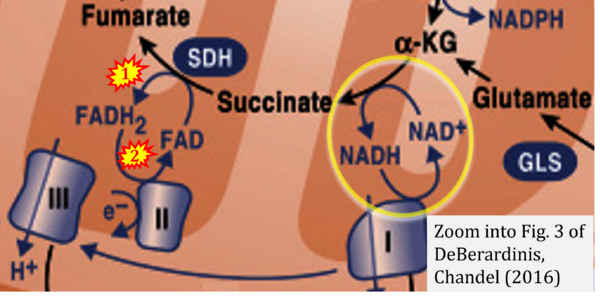
- DeBerardinis RJ, Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2:e1600200. - »Bioblast link«
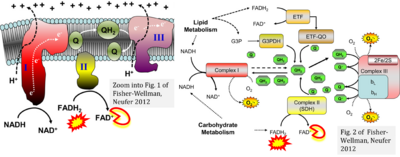
- Fisher-Wellman KH, Neufer PD (2012) Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab 23:142-53. - »Bioblast link«
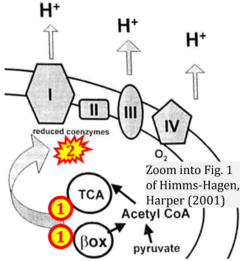
- Himms-Hagen J, Harper ME (2001) Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp Biol Med (Maywood) 226:78-84. - »Bioblast link«
- Sanchez H, Zoll J, Bigard X, Veksler V, Mettauer B, Lampert E, Lonsdorfer J, Ventura-Clapier R (2001) Effect of cyclosporin A and its vehicle on cardiac and skeletal muscle mitochondria: relationship to efficacy of the respiratory chain. Br J Pharmacol 133:781-8. - »Bioblast link«
- Oxidative phosphorylation by OpenStax Biology (CC BY 3.0) got it wrong in figures and text, and the error is propagated further, with copies in
- Conduct Science:"In Complex II, the enzyme succinate dehydrogenase in the inner mitochondrial membrane reduce FADH2 to FAD+. Simultaneously, succinate, an intermediate in the Krebs cycle, is oxidized to fumarate." - Comments: FAD does not have a postive charge. FADH2 is the reduced form, it is not reduced. And again: In CII, FAD is reduced to FADH2.
CII and fatty acid oxidation
- CII is not involved in fatty acid oxidation, which requires electron transferring flavoprotein CETF and CI. But there are confusing accounts of a rote of CII in fatty acid oxidation, which require correction:
- "Since mitochondrial Complex II also participates in the oxidation of fatty acids (6), .." (quote from [ Lemmi et al 1990]).
- Ref 6: Tzagoloff A (1982) Mitochondria. Plenum, New York.
- CII is not involved in fatty acid oxidation, which requires electron transferring flavoprotein CETF and CI. But there are confusing accounts of a rote of CII in fatty acid oxidation, which require correction:
- CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna - Acyl-CoA dehydrogenase is listed under 'Electron transfer in Complex II'.
MitoPedia concepts:
MiP concept
MitoPedia topics:
Enzyme,
Substrate and metabolite
Labels:
MitoPedia:FAT4BRAIN