The printable version is no longer supported and may have rendering errors. Please update your browser bookmarks and please use the default browser print function instead.
Description
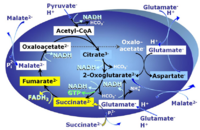
PGMS: Pyruvate & Glutamate & Malate & Succinate.
MitoPathway control state: NS-pathway control state
2-oxoglutarate is produced through the citric acid cycle from citrate by isocitrate dehydrogenase, from oxaloacetate and glutamate by the transaminase, and from glutamate by the glutamate dehydrogenase. If the 2-oxoglutarate carrier does not outcompete these sources of 2-oxoglutarate, then the TCA cycle operates in full circle with external pyruvate&malate&glutamate&succinate
Abbreviation: PGMS
Reference: Gnaiger 2020 BEC MitoPathways
PGMSL
- PGMS pathway in the LEAK state can be evaluated in the following SUIT protocols:
PGMSP
- PGMS pathway in the OXPHOS state can be evaluated in the following SUIT protocols:
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
- DL-Protocol for permeabilized cells (pce): SUIT-008 O2 ce-pce D025
PGMSE
- PGMS pathway in the ET state can be evaluated in the following SUIT protocols:
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-001 O2 mt D001
- DL-Protocol for permeabilized fibers (pfi): SUIT-001 O2 pfi D002
- DL-Protocol for permeabilized cells (pce): SUIT-001 O2 ce-pce D003
- DL-Protocol for permeabilized PBMC and PLT: SUIT-001 O2 ce-pce D004
- DL-Protocol for permeabilized fibers (pfi): SUIT-008 O2 pfi D014
- DL-Protocol for permeabilized cells (pce): SUIT-008 O2 ce-pce D025
Discussion
- Recent studies showed that S- and NS-linked OXPHOS capacity is inhibited by 2 mM malate concentrations as applied in many SUIT protocols. This inhibition is less pronounced at higher succinate concentrations (10 mM up to 50 mM S).
MitoPedia concepts:
SUIT state