Yusoff 2015 InTech
| Yusoff AAM, Ahmad F, Idris Z, Jaafar H, Abdullah JM (2015) Understanding mitochondrial DNA in brain tumorigenesis. In: Lichtor T, ed. Molecular considerations and evolving surgical management issues in the treatment of patients with a brain tumor. InTech: http://dx.doi.org/10.5772/58965 |
Yusoff AAM, Ahmad F, Idris Z, Jaafar H, Abdullah JM (2015) InTech
Abstract: In developed countries, most studies reveal that the number of people who develop brain tumors and die from them has increased. Brain tumor, one of the most devastating central nervous system pathologies, is the leading cause of solid tumor death in children under the age of 15, and the second leading cause of cancer death in male adults ages 20-39. So far, researches on genesis and development of tumor are intensively focused and studied on alteration of the gene in nucleus and brain tumors is the one where most were reported arise as the result of progressive nuclear genetic alterations. Multiple genetic events have been identified in brain tumor cells involving some well-known susceptibility genes such as tumor suppressor and oncogenes that are encoded by the nuclear DNA (nDNA). For instance, the p53 tumor suppressor gene is frequently mutated and often detected altered or lost early in brain tumor mainly in astrocytic tumors formation [1-3]. Similarly, mutations or loss of PTEN (phosphatase and tensin homolog), p16, RB (retinoblastoma) and amplification of EGFR (epidermal growth factor receptor), MDM2, CDK4, CDK6 (cyclin-dependent kinase) are also involved in the pathogenesis of brain tumor [4,5].
• Bioblast editor: Gnaiger E
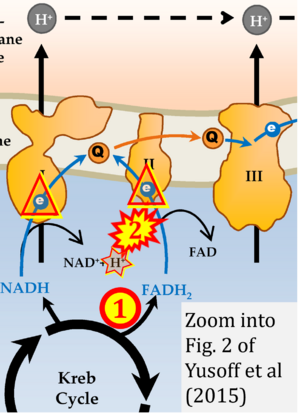
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
Labels:
Pathology: Cancer
Organism: Human Tissue;cell: Nervous system
Enzyme: Complex II;succinate dehydrogenase