Solhaug 2023 Cytotechnology
| Solhaug A, Gjessing M, Sandvik M, Eriksen GS (2023) The gill epithelial cell lines RTgill-W1, from Rainbow trout and ASG-10, from Atlantic salmon, exert different toxicity profiles towards rotenone. Cytotechnology 75:63-75. https://doi.org/10.1007/s10616-022-00560-0 |
Solhaug A, Gjessing M, Sandvik M, Eriksen GS (2023) Cytotechnology
Abstract: In order to ensure the proper use and interpretation of results from laboratory test systems, it is important to know the characteristics of your test system. Here we compare mitochondria and the handling of reactive oxygen species (ROS) in two gill epithelial cell lines, the well-known RTgill-W1 cell line from Rainbow trout and the newly established ASG-10 cell line from Atlantic salmon. Rotenone was used to trigger ROS production. Rotenone reduced metabolic activity and induced cell death in both cell lines, with RTgill-W1 far more sensitive than ASG-10. In untreated cells, the mitochondria appear to be more fragmented in RTgill-W1 cells compared to ASG-10 cells. Furthermore, rotenone induced mitochondrial fragmentation, reduced mitochondria membrane potential (Δψm) and increased ROS generation in both cell lines. Glutathione (GSH) and catalase is important to maintain the cellular oxidative balance by eliminating hydrogen peroxide (H2O2). In response to rotenone, both GSH and catalase depletion were observed in the RTgill-W1 cells. In contrast, no changes were found in the GSH levels in ASG-10, while the catalase activity was increased. In summary, the two salmonid gill cell lines have different tolerance towards ROS, probably caused by differences in mitochondrial status as well as in GSH and catalase activities. This should be taken into consideration with the selection of experimental model and interpretation of results.
• Bioblast editor: Gnaiger E
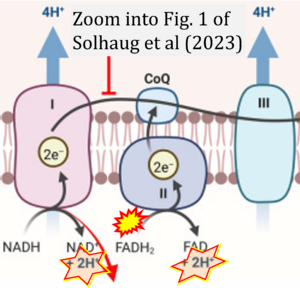
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.