Lu 2023 Explor Res Hypothesis Med
| Lu F (2023) Hypothetical hydrogenase activity of human mitochondrial Complex I and its role in preventing cancer transformation. Explor Res Hypothesis Med 8:280-5. https://doi.org/10.14218/ERHM.2022.00083 |
Lu Fuxiong (2023) Explor Res Hypothesis Med
Abstract: Cancer research has made a magnificent progress in past decades with an advancement of molecular biology. However, the mechanisms of cancer transformation are still not fully revealed. Thus, we must think about if there are some unknown factors playing a causative role in the cancer formation. Mitochondrial complex I oxidizes NADH to NAD+ and reduces ubiquinone to ubiquinol, regenerated NAD+ keeping pyruvate dehydrogenase and Krebs cycle function. Hydrogenases are widespread in nature, they occur in bacteria, archaea, and some eukarya. It is unknown whether hydrogenase activity exists in human mitochondria. The complex I shares a last common ancestor with hydrogenases, and is closely related with hydrogenase in sequence and modular structure. The hydrogenase activity has been observed recently in complex I of higher plants. Based on these observations, I propose a hypothesis that mitochondrial complex I in human may also retain the hydrogenase activity. The hypothetical hydrogenase activity could release excessive reducing equivalents of NADH from electron transport chain when a cell is in hypoxia, decreased oxidative phosphorylation or a low ATP demand. Loss of the hydrogenase activity may result in aerobic glycolysis, activation of pentose phosphate pathway, elevated lipid synthesis, and activations of oncoproteins via acetylation, all of these alterations lead to cell proliferations and cancer transformation. Reducing mitochondrial NADH/NAD+ ratio or recovering the hydrogenase activity would reverse the cell transformation.
• Bioblast editor: Gnaiger E
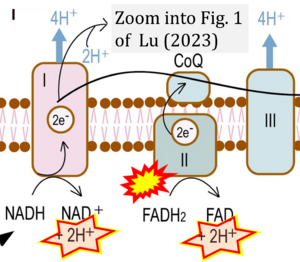
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
Labels: Pathology: Cancer