Joshi 2022 Biomolecules
| Joshi A, Ito T, Picard D, Neckers L (2022) The mitochondrial HSP90 paralog TRAP1: structural dynamics, interactome, role in metabolic regulation, and inhibitors. Biomolecules 12:880. https://doi.org/10.3390/biom12070880 |
Joshi A, Ito T, Picard D, Neckers L (2022) Biomolecules
Abstract: The HSP90 paralog TRAP1 was discovered more than 20 years ago; yet, a detailed understanding of the function of this mitochondrial molecular chaperone remains elusive. The dispensable nature of TRAP1 in vitro and in vivo further complicates an understanding of its role in mitochondrial biology. TRAP1 is more homologous to the bacterial HSP90, HtpG, than to eukaryotic HSP90. Lacking co-chaperones, the unique structural features of TRAP1 likely regulate its temperature-sensitive ATPase activity and shed light on the alternative mechanisms driving the chaperone's nucleotide-dependent cycle in a defined environment whose physiological temperature approaches 50 °C. TRAP1 appears to be an important bioregulator of mitochondrial respiration, mediating the balance between oxidative phosphorylation and glycolysis, while at the same time promoting mitochondrial homeostasis and displaying cytoprotective activity. Inactivation/loss of TRAP1 has been observed in several neurodegenerative diseases while TRAP1 expression is reported to be elevated in multiple cancers and, as with HSP90, evidence of addiction to TRAP1 has been observed. In this review, we summarize what is currently known about this unique HSP90 paralog and why a better understanding of TRAP1 structure, function, and regulation is likely to enhance our understanding of the mechanistic basis of mitochondrial homeostasis.
• Bioblast editor: Gnaiger E
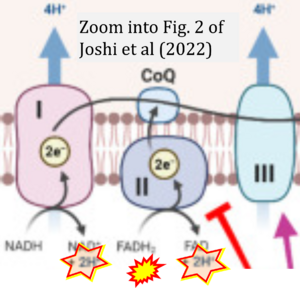
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
Labels:
Enzyme: Complex II;succinate dehydrogenase