From Bioblast
MitoFit-KMP
- The project MitoFit highlights the benefits of mitochondrial fitness.
WP2
- MitoFit-Knowledge Management Platform
Abstract
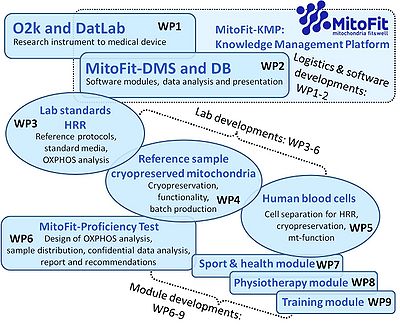
- Our goal is to establish an integration and analysis environment for measurements of mitochondrial respiration (O2k-technology) with external data (cell physiology, anthropometry, spiroergometry, lifestyle, genomics) in the context of fitness evaluation and clinical diagnostics. Within the scope of the MitoFit project, a Knowledge Management Platform (MitoFit-KMP) will be established. This KMP will accomplish the integration of all relevant data and provide a presentation platform for mitochondrial functional diagnostics. The MitoFit-KMP will include: a data management system for the current and planned instruments (MitoFit-DMS), and a database (MitoFit-DB) comprising proprietary and publicly available data that allow researchers to query experimental data and perform integrative analyses. Closely linked to WP1, a strategy will be developed on the transition of the DatLab software from a continuously updated and extended research tool to a component of the O2k as a medical device.
MitoFit-Knowledge Management Platform (KMP) with integrated MitoFit-Data Management System (DMS) and MitoFit-Data Bank (DB), which can be populated by external data related to the sample for respirometric measurement. The MitoFit-KMP will be an integral part of the O2k-technology, for data analysis tightly coupled to DatLab (the real-time software for the O2k), designed to export data into a MitoEAGLE Databank (Open Access, outside of the scope of the K-Regio project MitoFit).
Progress and next steps
- 2016 Jan - 2017 Jan: Christina Plattner, Dipl.-Ing. (Bioinformatics, Medical University Innsbruck)
- » Gnaiger 2016 Abstract EBEC Riva del Garda 2016: MitoFit and MitoEAGLE – towards a global data bank on mitochondrial physiology.
- » Garcia-Roves 2016a Abstract MitoFit Science Camp 2016: Design and implementation of systems biology approaches to integrate heterogenic data in biomedical research.